The so called collective behavior such as yawning might be an urban myth but a recent studies have attempted to model such collective behavior. The modal system they considered consists of an example of collective behavior of cows. Such theories have not been tested it in the real cows, however one should not be surprised to see such studies to be verified in near future.
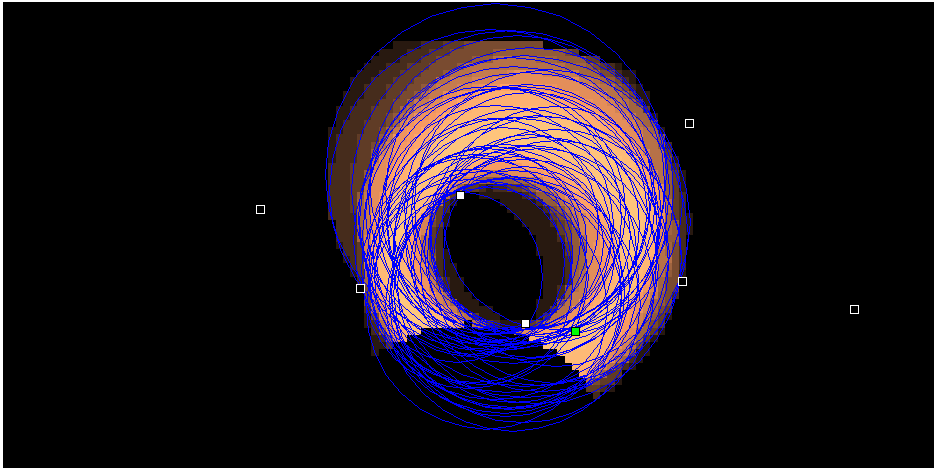
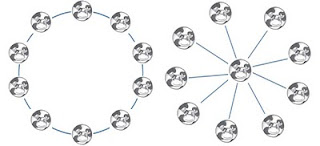
In a recent work, published in arxiv, Sun et. al. study the collective behavior of animals such as cows. Animals are coupled oscillators. They simply model the cow as coupled oscillator. By considering the discrete states of such animals and by considering such couplings, they study the collective decision bearing of such system.
Specifically, they consider three states of cows: standing, sitting and grazing; say "1", "2" and "3" just for sake of easy symbolization. By coupling the states, they assume that behavior of one is going affect the behavior of nearby ones. So, there is more tendency of uniform state such as 1,1,1,1,1 than 1,2,3,2,1 or 1,2,1,2,3. However, for larger cow population there can be some nice oscillatory behavior between the stable states.
The paper is interesting! Have a look.
Ref:
http://arxiv.org/abs/1005.1381
http://www.technologyreview.
Acknowledgements are due to the first author for suggests in the draft version of the post and allowing to use the figures.
On the lighter side:
One can see the influence of "spherical cow" on the coupling diagrams. See Jackson, J.D. Third Edition Ch3 problem#15.